De Novo Segmentation plugins

Cellpose-Predict plugin
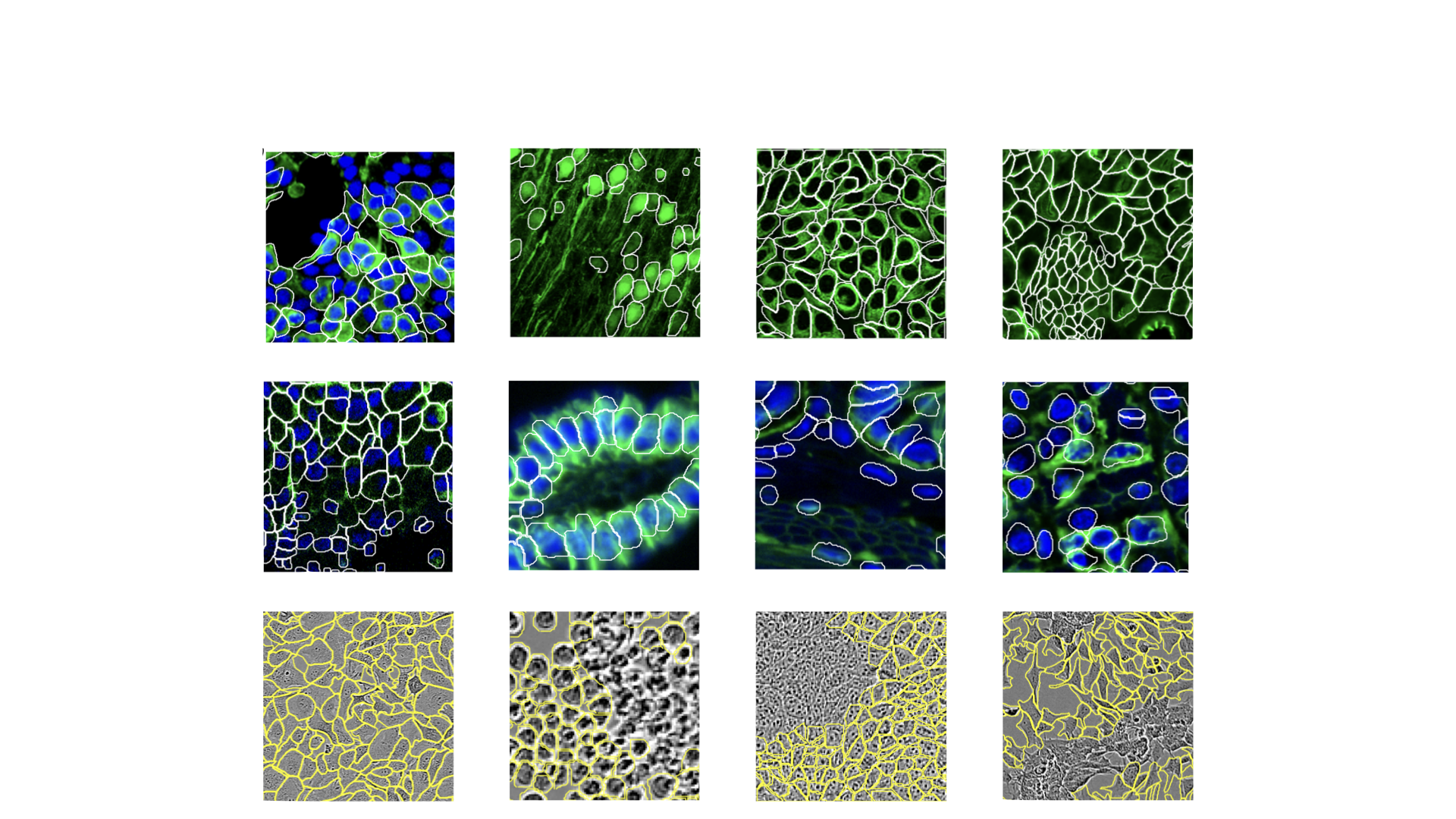
- class morphonet.plugins.DeNovoSegmentation.CellposePredict.CellposePredict
Bases:
MorphoPluginThis plugin uses an intensity image of the membranes from a local dataset at a specific time point, to compute a segmentation of the membranes, using the 3D CellPose deep learning algorithm. Users can apply CellPose on a 3D selected Mask to apply it to a part of a segmentation. By default, the plugin also disconnects all non-connex labels in the generated segmentation, and deletes all segmentations below a certain volume (in voxels), the same way it does in the DISCO and DELI plugins. Each of these operations can be disabled with the plugin parameters.
This plugin can be used:
on a whole image by not selecting any object.
on a sub-part of the image by selecting objects. The algorithm will then be applied only on the mask of these segmented objects
This plugin can use the base models provided by cellpose (default_model parameter), or a custom-made model (pretrained_model parameter), that can be created for instance by using the Cellpose-Train plugin.
- Parameters:
intensity_channel (int, default: 0) – in case of multiples channel, this corresponds to the intensity images channel
dimension (string) – to applied CellPose in 2D or 3D
time_points (string) – on which time point to train (current, begin:end, time1;time2;…)
downsampling (int, default: 2) – The resolution reduction applied to each axis of the input image before performing segmentation, 1 meaning that no reduction is applied. Increasing the reduction factor may reduce segmentation quality
default_model (string) – The model used to compute segmentations. A detailed documentation on models can be found https://cellpose.readthedocs.io/en/latest/models.html
pretrained_model (string) – the full filename path to for pretrained model
diameter (int, default: 30) – the average cell size diameter (in voxels)
isotrope (bool, default: True) – resize (or not) the image to be isotropic before making prediction
objects (Disconnect) – split any object made up of several sub-objects that are not in contact with each other.
volume (Minimum) – reassign id with small objects under the value to their closet neigbhors. 0 will not perform any reassignment
Reference –
--------- – Stringer, C., Wang, T., Michaelos, M. et al. Cellpose: a generalist algorithm for cellular segmentation. Nat Methods 18, 100?106 (2021). https://doi.org/10.1038/s41592-020-01018-x https://www.cellpose.org

Cellpose-Train plugin
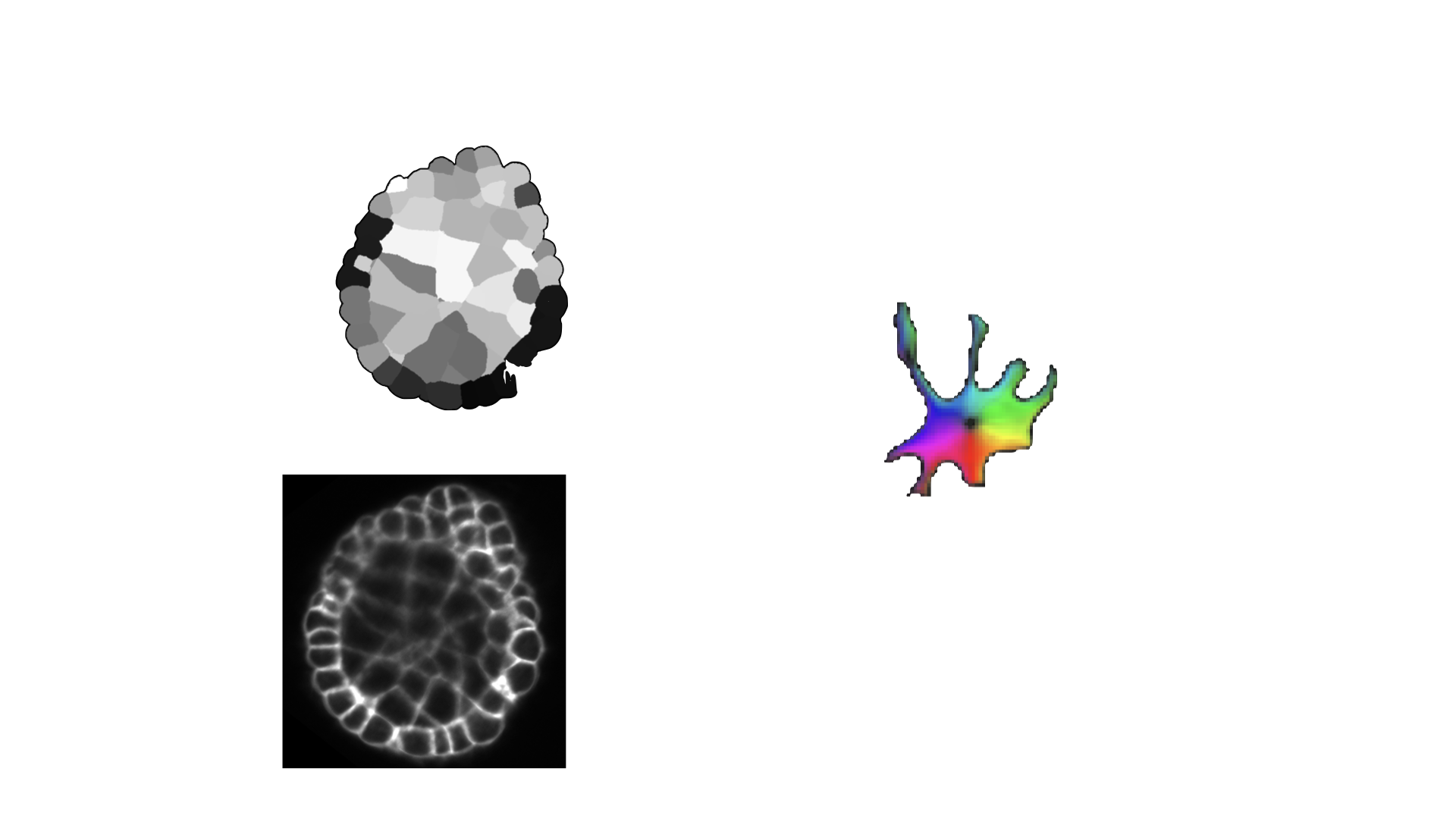
- class morphonet.plugins.DeNovoSegmentation.CellPoseTrain.CellposeTrain
Bases:
MorphoPluginThis plugin allows users to train their own model (from the models provided by CellPose) on their own 3D datasets. With an intensity image and the corresponding segmentation, you can re-train a cellpose model to obtain your own model, trained on your images. The model will be trained by using 2D images, which are the image stacks on the XY, XZ and YZ planes of your 3D images.
You can use this plugin:
on the whole image by not selecting any object.
on a sub-part of the image by selecting objects. The algorithm will then run on the masks of ALL the segmented objects found in the bounding box containing all the selected objects.
The model you output from this plugin can then be used in the Cellpose prediction plugin, by inputting it in the pretrained_model parameter.
- Parameters:
intensity_channel (int, default : 0) – in case of multiples channel, this corresponds to the intensity images channel
epochs (int, default : 20) – number of epochs of training
dimension (string) – For 2D: use XY planes to train. For 3D: extract XY, YZ,YX planes to train
time_points (string) – on which time point to train (current, begin:end, time1;time2;…)
number_2D_images_per_epochs (int, default : 8) – batch size
downsampling (int, default :2) – The resolution reduction applied to each axis of the input image before performing segmentation, 1 meaning that no reduction is applied. Increasing the reduction factor may reduce segmentation quality
model_type (string) – The model used to compute segmentations. A detailed documentation on models can be found https://cellpose.readthedocs.io/en/latest/models.html
model_filename (string) – full filename path to save the model
isotrope (bool, default: True) – resize (or not) the image to be isotropic before training
Reference –
--------- – Stringer, C., Wang, T., Michaelos, M. et al. Cellpose: a generalist algorithm for cellular segmentation. Nat Methods 18, 100?106 (2021). https://doi.org/10.1038/s41592-020-01018-x https://www.cellpose.org

Mars plugin
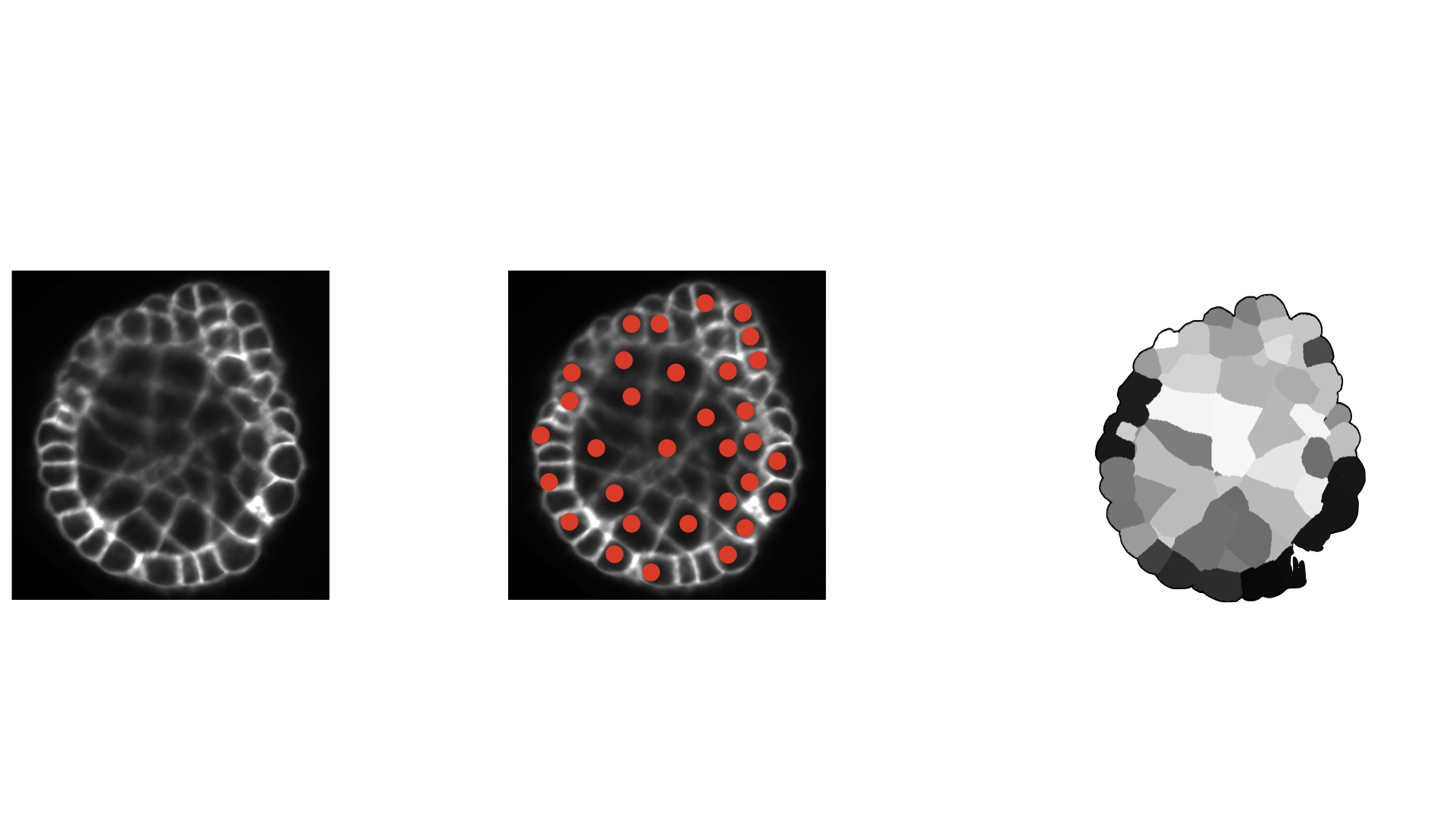
- class morphonet.plugins.DeNovoSegmentation.Mars.Mars
Bases:
MorphoPluginThis plugin uses an intensity image from a local dataset at a specific time point, to perform a segmentation using a seeded watershed algorithm.
This plugin requires intensity image(s), and is applied on the whole image.
- Parameters:
downsampling (int, default: 2) – The resolution reduction applied to each axis of the input image before performing segmentation, 1 meaning that no reduction is applied. Increasing the reduction factor may reduce segmentation quality (>=1)
sigma (int, default: 8) – The standard deviation for the Gaussian kernel when sigma>0, the plugin first applies a gaussian filter on the intensity image. (>=0)
h_minima_value (int, default: 2) – If using the H Minima method, determines the minimal depth of all extracted minima. Optional , >= 0
method (string) – Method used to find local minima of intensity in the intensity image. H Minima method determine all minima of the image with depth >= h_minima_value. Local minima method finds the local minima in the intensity image. The local minima are defined as connected sets of voxels with equal gray level (plateaus) strictly smaller than the gray levels of all voxels in the neighborhood.
membrane_channel (int , default: 0) – The desired channel to apply the segmentation
Reference
Fernandez R, Das P, Mirabet V, Moscardi E, Traas J, Verdeil JL, Malandain G, Godin C. Imaging plant growth in 4D: robust tissue reconstruction and lineaging at cell resolution. Nat Methods. 2010 Jul;7(7):547-53. doi: 10.1038/nmeth.1472. Epub 2010 Jun 13. PMID: 20543845.

Stardist-Predict plugin
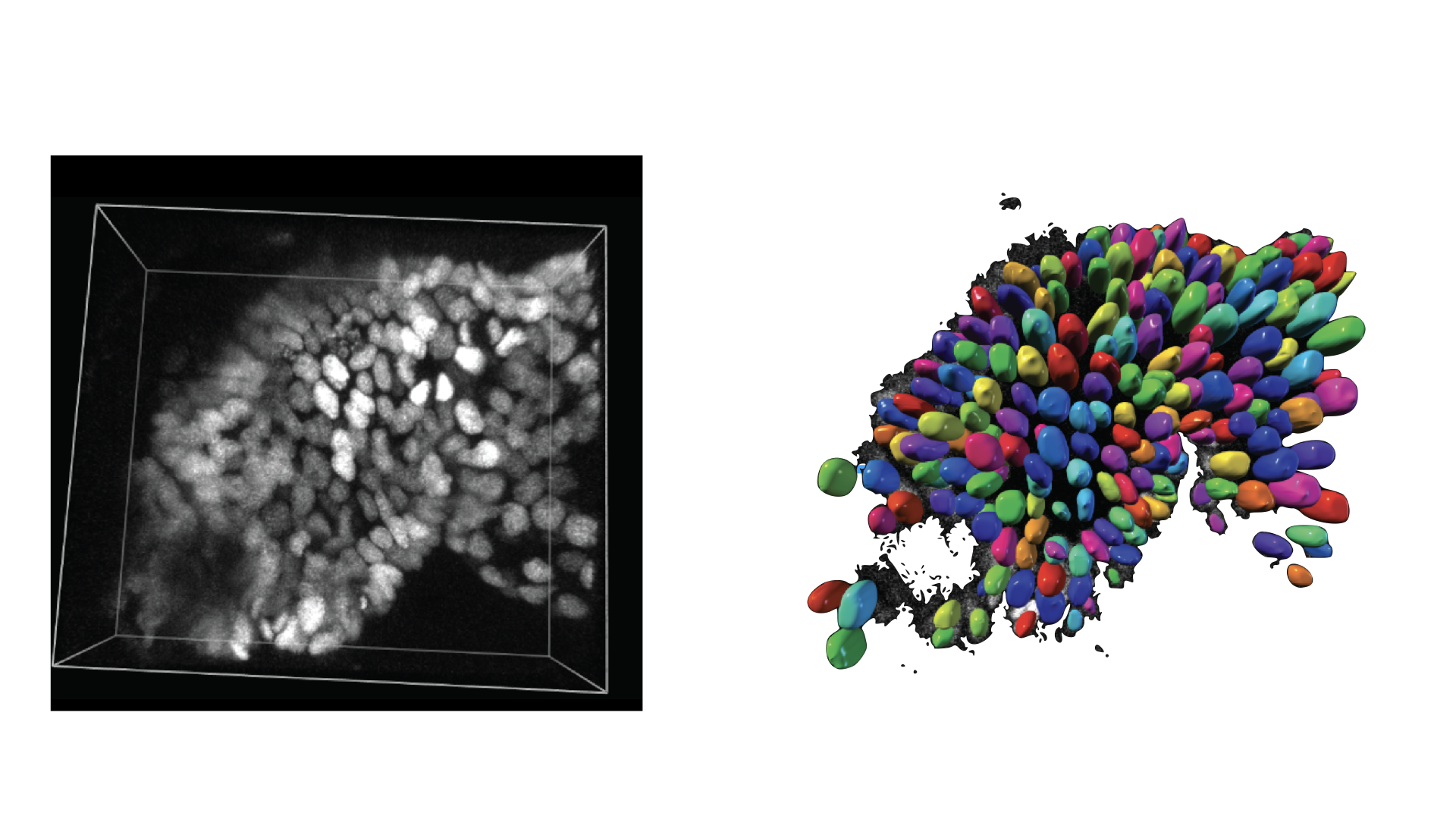
- class morphonet.plugins.DeNovoSegmentation.StardistPredict.StardistPredict
Bases:
MorphoPluginThis plugin uses an intensity image of the nuclei from a local dataset, to compute segmentations of the nuclei, using the 3D Stardist deep learning algorithm. The default demo model of stardist can be used, as well as custom models (that can be created by the Stardist-Train plugin).
This plugin requires intensity image(s), and is applied on the whole image.
- Parameters:
downsampling (int, default :2) – The resolution reduction applied to each axis of the input image before performing segmentation, 1 meaning that no reduction is applied. Increasing the reduction factor may reduce segmentation quality
nms_thresh (float, default :0.3) – Non-max suppression threshold in Stardist
nuclei_channel (int, default :0) – The desired channel to run stardist on
prob_thresh (float, default :0.707933) – Mask probability threshold in Stardist
time_points (string, default: current) – The time range for stardist
use_tiles (bool, default :True) – Should divide the process in tiles, recommended for big images
Pretrained_model (string) – the model filename (ex : weights_best.h5) used for the prediction (Default 3D model ) if the thrshod file isavialble in the folder nms_thresh and prob_thresh will be direcly extract from it
Reference –
--------- – Uwe Schmidt, Martin Weigert, Coleman Broaddus, and Gene Myers. Cell Detection with Star-convex Polygons. International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Granada, Spain, September 2018. https://github.com/stardist/stardist

Stardist-Train plugin
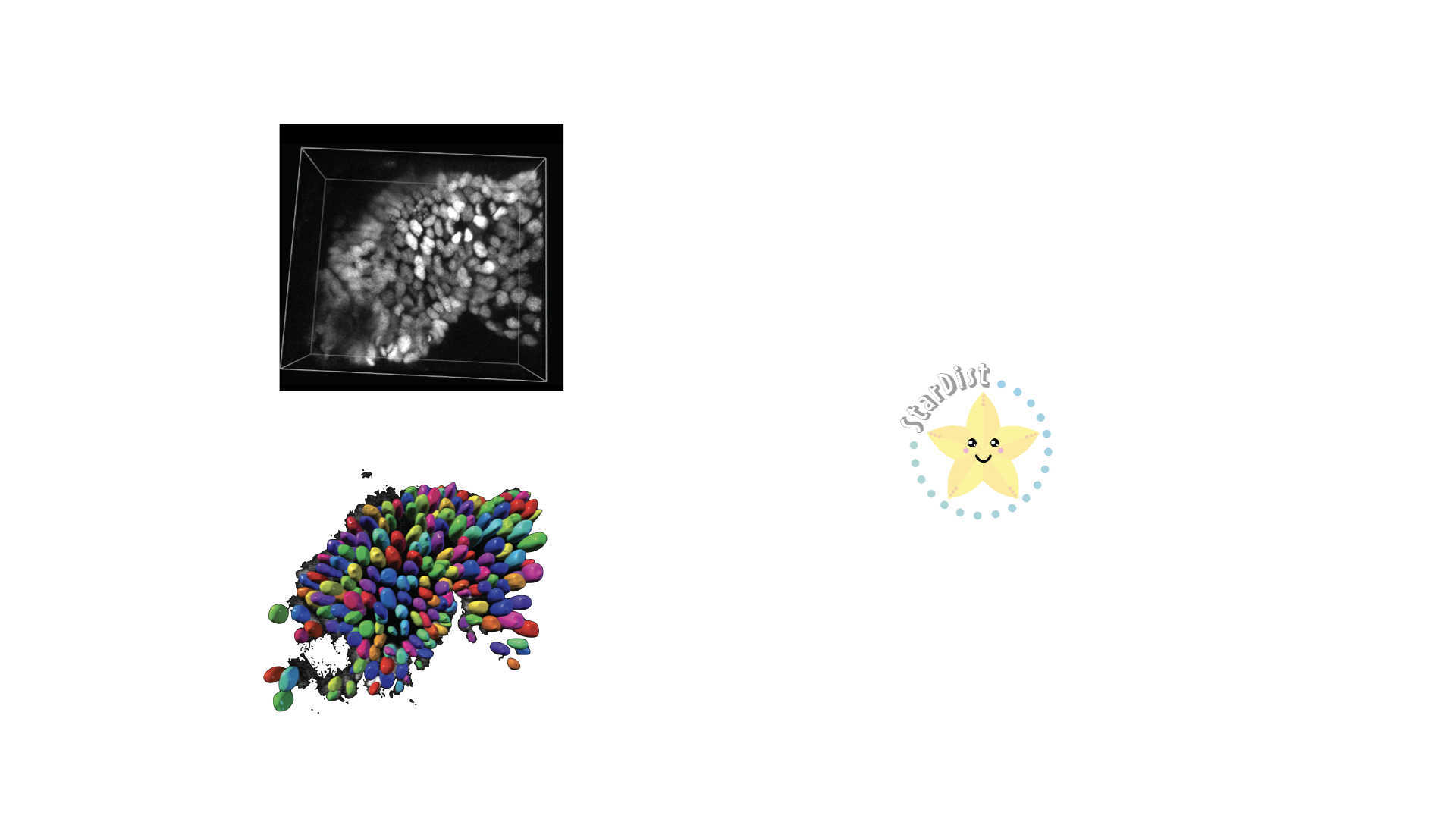
- class morphonet.plugins.DeNovoSegmentation.StardistTrain.StardistTrain
Bases:
MorphoPluginThis plugin allows users to train their own Stardist model on their own 3D datasets. Using the 3D intensity image with the corresponding segmentation, you can train your own Stardist model on several time points. Then the model can be used with the StardistPredict plugin to perform nuclei prediction on 3D intensity images.
This plugin requires intensity image(s), and is applied on the whole image.
- Parameters:
downsampling (int, default :2) – The resolution reduction applied to each axis of the input image before performing segmentation, 1 meaning that no reduction is applied. Increasing the reduction factor may reduce segmentation quality
nuclei_intensity_channel (int, default :0) – The desired channel to run stardist on
epochs (int, default :20) – Number of epochs to train the model
batch_size (int, default :2) – size of the batch size (increase learning process, but can create memory errors)
time_points (string, default: current) – The time range to train your model
model_filename (string) – full filename path to save the model
Reference –
--------- – Uwe Schmidt, Martin Weigert, Coleman Broaddus, and Gene Myers. Cell Detection with Star-convex Polygons. International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Granada, Spain, September 2018. https://github.com/stardist/stardist
- morphonet.plugins.DeNovoSegmentation.StardistTrain.augmenter(x, y)
Augmentation of a single input/label image pair. x is an input image y is the corresponding ground-truth label image

Binarize plugin
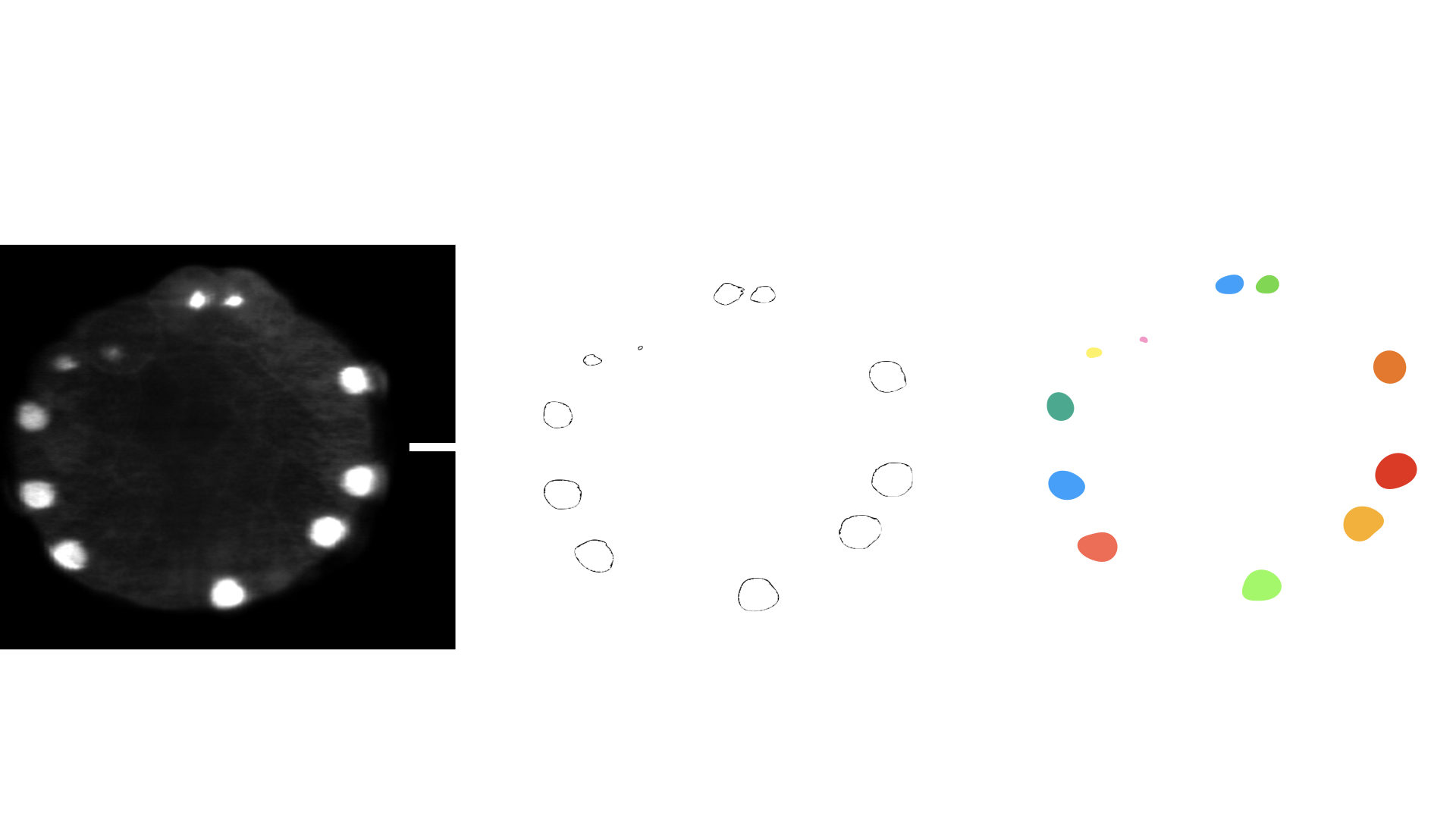
- class morphonet.plugins.DeNovoSegmentation.Binarize.Binarize
Bases:
MorphoPluginThis plugin applies a binary threshold to an intensity image and creates labels on each connected component above the threshold.
This plugin can be used:
on a whole image by not selecting any object.
on a sub-part of the image by selecting objects. The algorithm then runs on the mask of the selected objects.
- Parameters:
threshold (int, default: 1) – The threshold value
time_points (string , default: current) – The time range for binarization
intensity_channel (int, default: 0) – The desired channel to apply the binary threshold
BinCha plugin
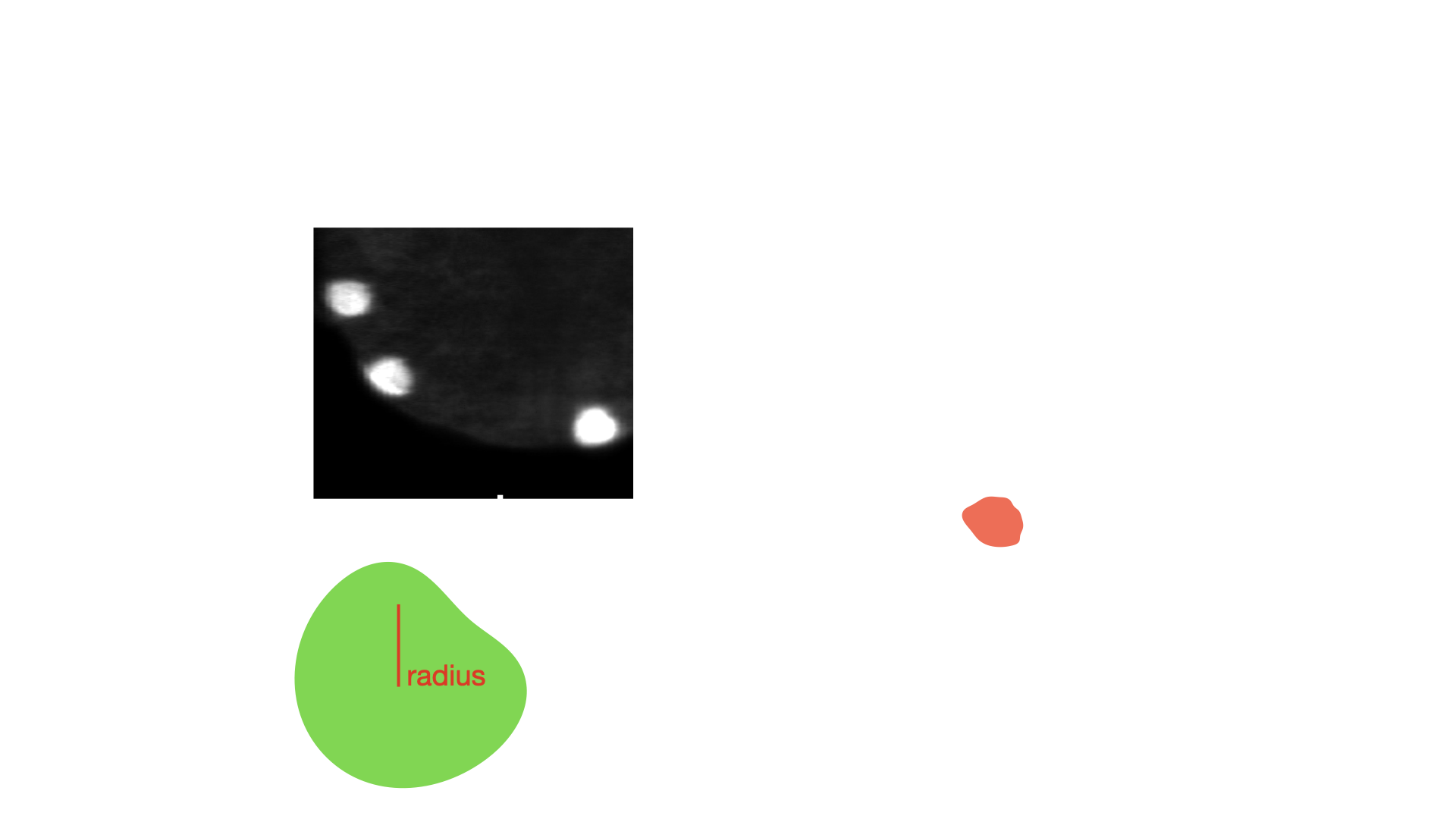
- class morphonet.plugins.DeNovoSegmentation.BinCha.BinCha
Bases:
MorphoPluginThis plugin creates a new segmentation from each mask of the selected objects using a binary threshold on intensity image in the desired channel. Alternatively, you can also do the thresholding on the centroid of each object, with a bounding box of input radius.
- Parameters:
intensity_Channel (int, default: 0) – The desired channel to apply the binary threshold
threshold (int, default: 30) – The threshold value using in the intensity image
radius (int, default: 15) – The radius value of the bounding box around the center of each selected objects (only applies is bbox_on_centroid is True)
intensity_channel (int, default: 0) – The desired channel to apply the binary threshold
bbox_on_centroid (bool, default: False) – Toggle on to compute on the centroid of selected objects, with a bounding box of radius

BinBox plugin
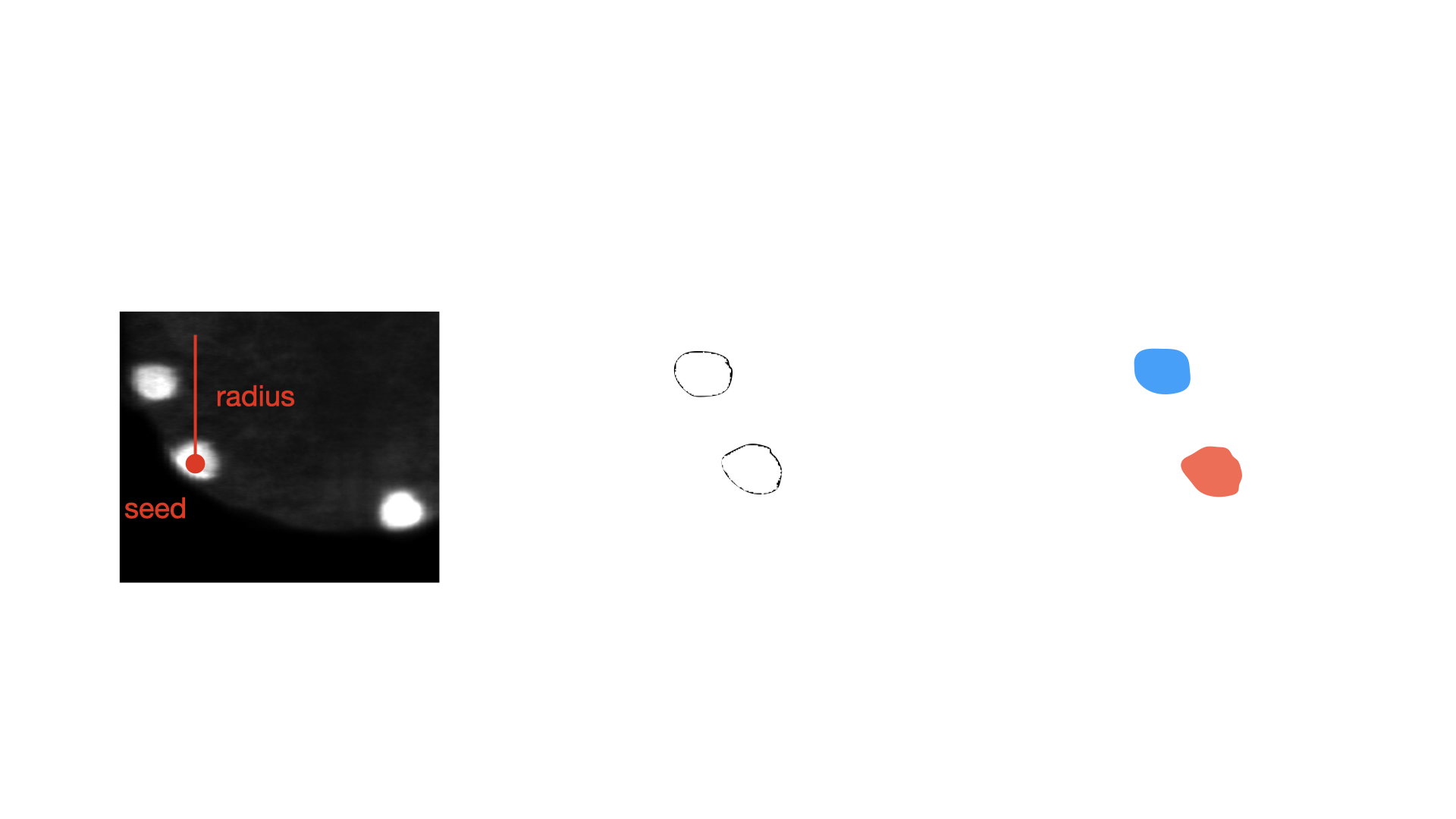
- class morphonet.plugins.DeNovoSegmentation.BinBox.BinBox
Bases:
MorphoPluginThis plugin applies a threshold on the intensity image on a bounding box (centered around a seed), and creates new labels on each connected component above the threshold.
This plugin requires at least one seed, as well as an intensity image.
- Parameters:
threshold (int, default: 1) – The threshold value
time_points (string, default: current) – The time range for binarization
radius (int, default : 15) – The radius value of the bounding box around the center of each selected objects
intensity_channel (int, default: 0) – The desired channel to apply the binary threshold
Seed – A seed object added in MorphoNet Interface